T. Vernet, C. Morlot, IBS : Morphogenesis of Streptococcus pneumoniae
Objectives
We study the cell wall synthesis mechanisms of the major human pathogen S. pneumoniae in order to understand how this ovoid bacterium grows, divides and acquires its shape; and to decipher its mechanisms of antibiotic resistance. In particular, the Gral project aims at developing the use of the super-resolution imaging method of PALM (PhotoActivated Localization Microscopy) in the pneumococcus.
Background
During growth and division, bacteria remodel the peptidoglycan (PG) of their cell wall. The PG, made of glycan chains cross-linked by peptide bridges, procures mechanical resistance and bacterial shape. Over the cell cycle, PG is bio-synthesized by multi-protein machineries (Fig. 1) located at the division site. They include the Penicillin-Binding Proteins (the PBPs, which are the targets of the beta-lactam antibiotics) for the polymerization of the glycan chains and their reticulation, as well as cell wall hydrolases for the controlled cleavage of the PG. Fine-tuning between synthesis and degradation relies on a complex network of interactions between these enzymes and other proteins with structural and/or regulatory functions. In addition, PG bio-synthesis is coordinated with membrane constriction, leaded by the tubulin homologue FtsZ.
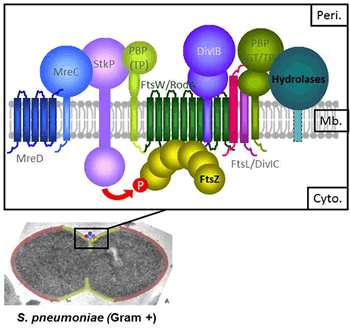
Figure 1. Growth and division proteins of S. pneumoniae. Composition of the machinery coupling cell wall synthesis, degradation and membrane constriction.
Team
Pneumococcus Group
- Thierry Vernet: group leader
- Cecile Morlot: CR1 CNRS
- Anne-Marie DI Guilmi: CEA research director
- André Zapun: CR1 CNRS
- Claire Durmort: Assistant professor (Maître de conference)
- Laure Bellard: CEA technicien
- Anne-Marie Villard: CEA technicien
Publications
Jacq M., Adam V., Bourgeois D., Di Guilmi A.M., Vernet T., Morlot C. New sub-structures of the FtsZ-ring from a pathogenic coccus revealed by super-resolution microscopy (PALM). Manuscript in preparation
Gral recruitment
Maxime Jacq (thesis started in October 2012)
M. Jacq is currently studying two aspects of pneumococcal morphogenesis. The first part of his Ph.D. is dedicated to developing the use of PhotoActivated Localization Microscopy (PALM) in S. pneumoniae, in collaboration with the Pixel team (V. Adam, D. Bourgeois, experts in fluorescent protein engineering and super-resolution microscopy) at the IBS. This super-resolution microscopy technique will allow getting fluorescence localization patterns at 20 nm resolution, thus revealing cellular details that were so far inaccessible with conventional microscopy. The second part of his Ph.D. focuses on the cell wall hydrolase Pmp23. This protein was discovered by the Pneumococcus Group as the first putative lytic transglycosylase. Its deletion results in abnormal shape, misplaced septa and increased sensitivity to beta-lactams. As the only identified transmembrane hydrolase, it could possibly interact with other morphogenetic membrane proteins. The objective of the Pmp23 project is to determine the role of the protein in PG bio-synthesis, a project that includes the study of Pmp23 cellular localization, enzymatic activity and protein interactome.
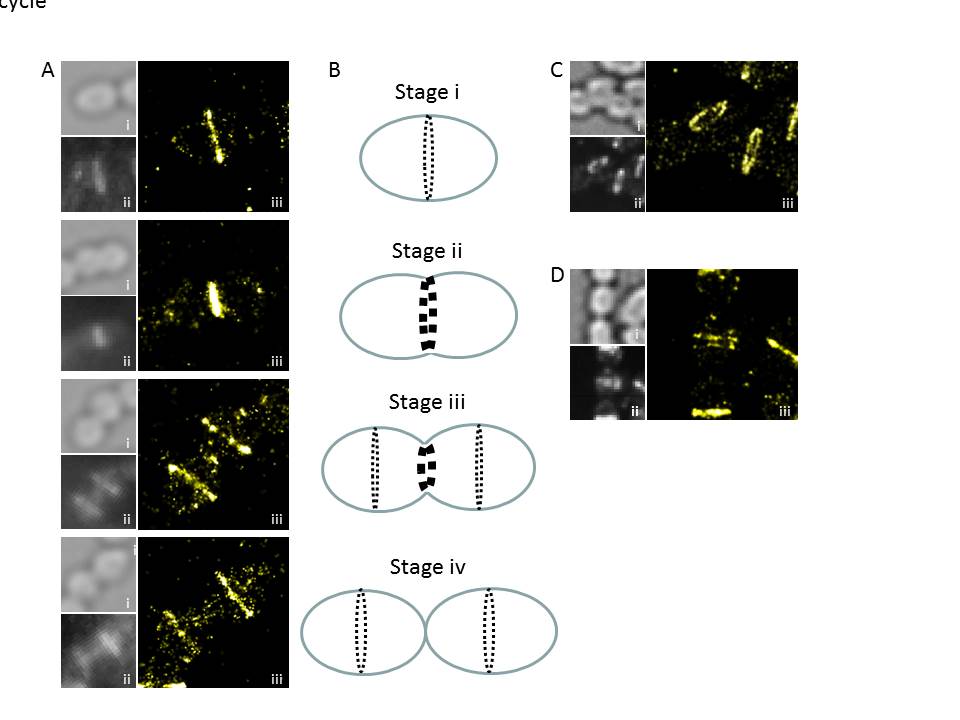
Figure 2. PALM nanostructure of FtsZ from S. pneumoniae. A. PALM patterns (iii) displayed by FtsZ along the cell cycle, together with bright field (i) and diffraction-limited (ii) images. B. Scheme the depicting the main stages in FtsZ localization during the division of the pneumococcus. C. FtsZ PALM pattern in tilted cells, showing the annular geometry of the FtsZ-ring. D. Observation of double rings of FtsZ, their function is currently under investigation.
