Deciphering assembly, function and structure of molecular nanomachines of the human pathogen Pseudomonas aeruginosa
Objectives
Study complex structures involved in bacterial secretion systems by using structural biology, microbiology, and cell biology techniques
Background
Both pathogenic and non-pathogenic bacteria deploy molecular weaponries to invade and colonize host eukaryotic cells or to compete with other bacteria within a microenvironment of the host organism. This project focuses on secretion systems (T2SS, T3SS, T6SS) of pathogens such as Pseudomonas aeruginosa. Understanding the regulation of these systems as well as function and characterization of the role of individual components of these molecular machineries is essential for the design of efficient therapeutics targeting virulence factors of a number of pathogens.
One of the goals of this project is to describe the function of novel – or yet uncharacterized – components of T3SS and T6SS apparatus of P. aeruginosa, including structural and regulatory components and secreted toxins, in order to better understand the underlying mechanisms of P. aeruginosa pathogenesis. Several new protein interactions have recently been identified in our laboratory within each of the two secretion systems using biochemistry and proteomics approach. However, the subcellular localization and function of these protein interactions in context of the (live) cell and kinetics of these interactions during physiological changes that occur during T3SS or T6SS activation remain largely unknown. To address this we are developing robust and quantitative microscopy-based imaging assays to analyze individual T3SS/T6SS components of P. aeruginosa on single-cell level and to describe dynamics of T3SS/T6SS gene expression and syringe assembly within entire bacterial populations. Electron microscopy and fluorescence time-lapse microscopy are being used followed by semi-automated image segmentation for quantitative analysis.
The second objective of this project involved the structural characterization of the apparatus itself. This effort counted on a collaboration with the electron microscopy group at the PSB (Schoehn group). In order to tackle this issue, we chose to study the secretin, the outer membrane ring-forming protein that is common to a number of secretion systems and plays key roles in cytotoxicity. In addition to being funded by GRAL, this work was also funded by an ANR project (SECRETIN) in collaboration with the Pasteur Institute.
Team
BIG : Bacterial Pathogenesis and Cellular Responses – Web: http://big.cea.fr/drf/big/BCI/PBRC
IBS :
- Andrea Dessen, research director and group leader (Bacterial Pathogenesis)
- Viviana Job, staff researcher
- Tommaso Tosi, postdoctoral fellow
- Guy Schoehn, research director and group leader (Microscopy)
- Leandro Estrozi, staff scientist
GRAL recruitments :
- Tommaso Tosi, postdoctoral fellow – IBS
- David Liebl, postdoctoral fellow – BIG
Publications
Tosi, T, Estrozi, LF, Job, V., Guilvout, I, Pugsley, AP, Schoehn, G, and Dessen, A (2014) Structural similarity of secretins from type II and type III secretion systems. Structure 22, 1348-1355.
Golovkine G., Faudry, E., Bouillot, S., Voulhoux, R., Attrée, I. and Huber, P. 2014. VE-Cadherin cleavage by LasB protease from Pseudomonas aeruginosa facilitates type III secretion system toxicity in endothelial cells. PLOS Pathog., 10 (3), e1003939.
Elsen**, S., Huber**, P., Bouillot, S., Couté, Y., Fournier, P., Dubois, Y., Timsit, JF., Maurin, M. and Attrée, I. 2014. A type III secretion negative clinical strain of Pseudomonas aeruginosa employs a two-partner secreted Exolysin to induce hemorrhagic pneumonia. Cell Host Microbe, 15(2):164-76. Spotlighted in Trends in Microbiology, Allen et al., 2014.
Achievements, part 1
- A collection of P. aeruginosa strains expressing selected components of T3SS and T6SS tagged with sfGFP or mCherry fluorescent proteins has been generated and the imaging protocol has been set up to analyze bacterial cells during (i) in vitro infection of the human epithelial cells (for T3SS); (ii) growth on semi-solid substrate in biofilm-like conditions; (iii) competition within mixed bacterial populations (for T6SS).
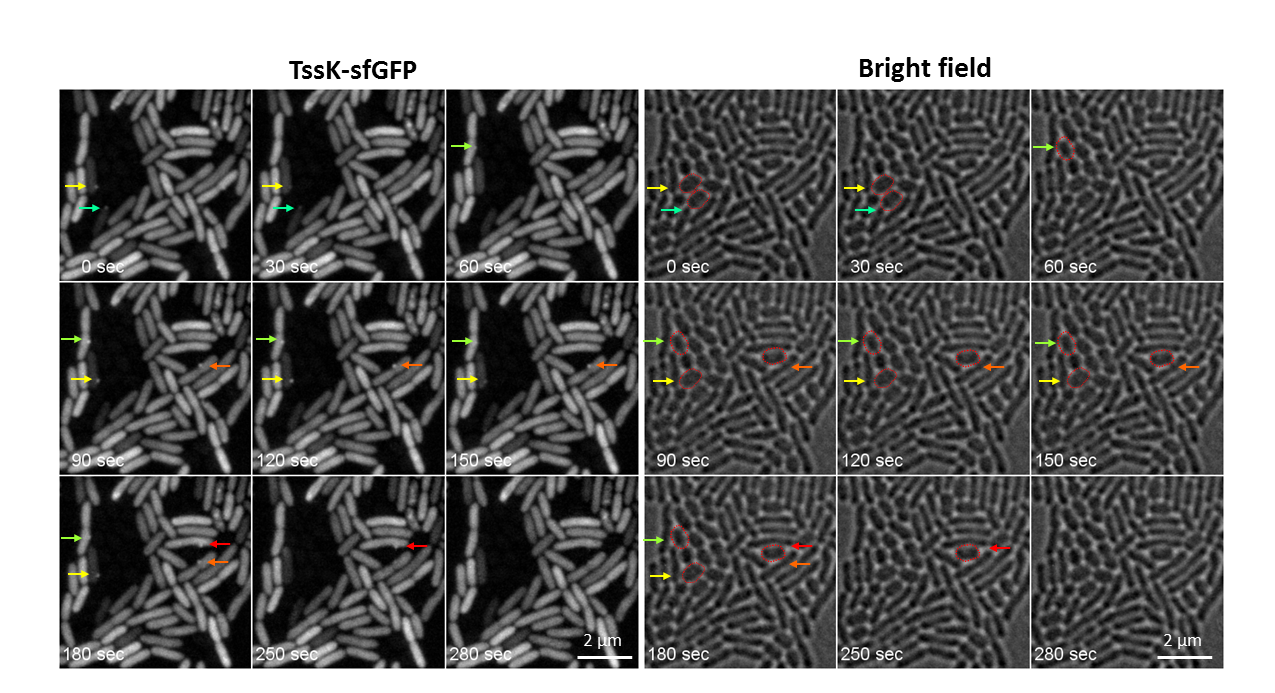
- We identified TssK as interacting component with other membrane components of T6SS machinery. We revealed that TssK localizes to cytosol but assembles into higher order structure which can be detected as a high intensity fluorescent spot(s) localizing predominantly to bacterial membrane. One to two TssK-sfGFP spots per cell were detected under inducing conditions. We found that assembly of TssK-positive structure(s) within bacterial cells is dependent on presence of T6SS signaling component TagQ and can be transiently induced in response to cell membrane damage. Time-lapse analysis of live cells revealed that TssK-spots are not dynamic and do not undergo frequent assembly-disassembly (as assessed by single-spot fluorescence density measurement). We thus propose that TssK – a putative base plate component – has a capacity to assemble from cytosolic pool into membrane-embedded structure to provide stabilized anchor serving as a spatio-temporal cue for assembly of the contractile syringe at the cytosolic side of the inner membrane of bacteria.
- To further investigate in which particular stage of T6SS machinery (activation/assembly/contraction/disassembly) TssK participates, we focused on spatio-temporal correlation between TssK-spots and highly dynamic assembly of contractile sheeth of the T6SS syringe composed of TssB. Cells expressing TssB-sfGFP fusion construct were monitored by time-lapse fluorescence microscopy and kinetics of assembly-disassembly has been described. Interestingly, time-lapse imaging of both, homogenous populations of P. aeruginosa and mixed cultures with A. baumanni revealed that deployment of T6SS apparatus occurs in random directions rather as a response to contact with a target cell. However, we also found that in context of a single cell the repeated assembly of TssB-GFP syringe often occurred at the same subcellular localization. This would imply that TssB polymerization initializes at a preexisting “priming site”, but whether these sites correspond to pre-assembled TssK structure remains to be confirmed in cells coexpressing TssB-GFP with TssK-TagRFP (in progress). The role of two other TssK-binding partners TssG and TssE, in T6SS assembly/deployment is under investigation.
- An image-based assay to screen for functional assembly of T3SS apparatus has been established using immunolabeling of PcrV, an extracellularly exposed substructure at the tip of the T3SS needle, as a read-out of T3SS assembly. This assay was used to evaluate panel of mutants to determine protein residues within PscI and PscF that are essential for secretion-capable assembly of the T3SS apparatus. Two point-mutations within PscI were found to completely abrogate needle assembly so as toxin secretion (confirmed by biochemical assay). To determine the impact of mutations on needle structure, needles will be further assessed by electron microscopy.
- Cell-free expression of secretins from P. aeruginosa and K. oxytoca allowed for the preparation of soluble, stable protein that generated excellent images in negative stain electron microscopy.
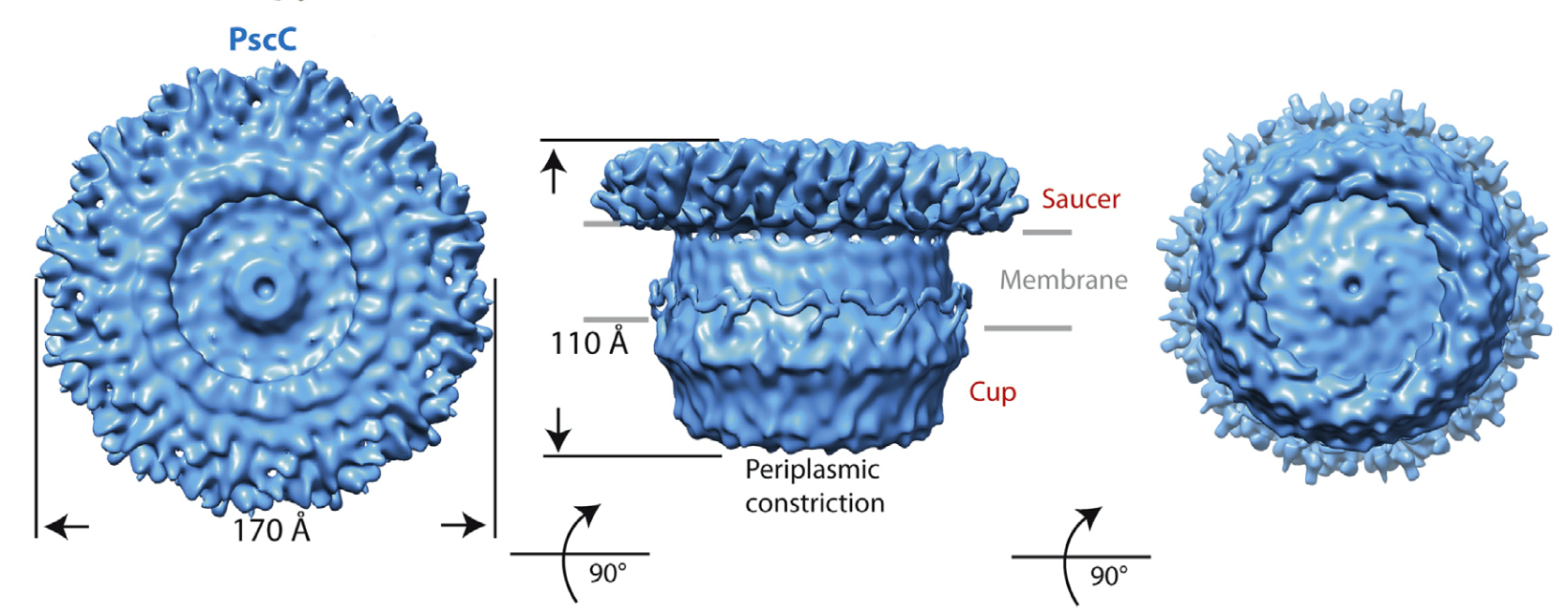
- Cryo electron microscopy of the samples allowed for the establishment that both secretins have a ‘cup and saucer’ architecture, with two outer rings forming the saucer region
- Detailed analyses of the samples allowed for the identification of an interior cavity within the structure of the saucer, as well as a ‘double wall’ architecture for the cup
- The two structures resemble the ‘closed’ forms of secretins previously described in the context of entire secretion systems.
Figure 1 : T6SS deployment in mixed bacterial population of PAO1 (wild type) and Acinetobacterbaumanni: Time-lapse imaging of live PAO1 cells expressing TssK-sfGFP under inducible promotor and grown for 1hr in mixed culture with Acinetobacterbaumanni. Selected frames are shown from a time-lapse imaging of live cells (TssK-sfGFP fluorescence on left, bright field on right), 30 sec between frames, Mag. 160x. Assembly of TssK-sfGFP spots in PAO1 cells occurs towards the contact with prey cell (A. baumanni). Assembly-disassembly of individual TssK-spots is indicated by color-coded arrows. Targeted prey cells of A. baumanni are encircled in red in corresponding bright field images (right panel)
Achievements, part 2
- Cell-free expression of secretins from P. aeruginosa and K. oxytoca allowed for the preparation of soluble, stable protein that generated excellent images in negative stain electron microscopy.
- Cryo electron microscopy of the samples allowed for the establishment that both secretins have a ‘cup and saucer’ architecture, with two outer rings forming the saucer region
- Detailed analyses of the samples allowed for the identification of an interior cavity within the structure of the saucer, as well as a ‘double wall’ architecture for the cup
- The two structures resemble the ‘closed’ forms of secretins previously described in the context of entire secretion systems.
Figure 2: Cryo EM structure of PscC, the secretin from the type III secretion system of the human pathogen Pseudomonas aeruginosa (Tosi, Estrozi et al (2014) Structure 22, 1348-1355.